Read counts are summarized for various region types and the corresponding aggregate count matrices are used for dimension reduction. Counts have been log-normalized (log10(count+1)).
This ATAC-seq dataset contains accessibility profiles for 142 samples.
Signal has been summarized for the following region sets:
| #regions | transformations | |
| t200 | 13140638 | quantileNorm |
| t10k | 273390 | quantileNorm |
| promoters_gc_protein_coding | 18791 | quantileNorm |
| ERB_BP_distal | 79617 | quantileNorm |
| ERB_BP_dnase | 69129 | quantileNorm |
| ERB_BP_proximal | 99810 | quantileNorm |
| ERB_BP_tfbs | 14111 | quantileNorm |
| ERB_BP_tss | 19985 | quantileNorm |
| .peaks.cons | 558050 | quantileNorm |
Fragment data IS available.
Read counts are summarized for various region types and the corresponding aggregate count matrices are used for dimension reduction. Counts have been log-normalized (log10(count+1)).
Samples have been clustered according to the 1000 most variable regions for each region type. Counts have been log-normalized (log2(count+1)).
| Region type |
Clustered heatmap. The 1000 most variable regions are shown. Counts have been log-normalized (log2(count+1)).
chromVAR [1] analysis. The following motif set(s) were used for the analysis: jaspar_vert. R data files of chromVAR deviation scores have been attached to this report:
| Region type |
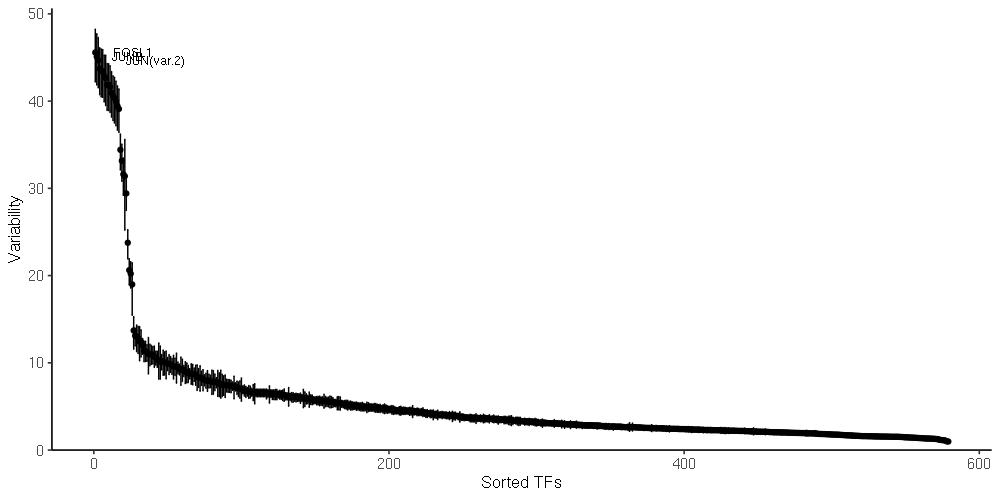
chromVAR variability. TF motifs are shown ordered according to their variability across the dataset.
| Region type |
chromVAR deviation scores. The heatmap shows the scores for 100 most variable TF motifs across the dataset.